Dissecting protein networks and regulatory mechanisms driving ciliary disassembly
Shibu Antony (ESR3)
Shibu is a PhD candidate in Experimental Medicine at the Medical Faculty of the University of Tübingen, working with the cilia group at the Institute for Ophthalmic Research. During his PhD he worked on studying the role of cilia in the context of cell cycle progression and generating a protein interaction network map for different regulators that have been previously reported to be involved in controlling the process of cilia disassembly. Through the course of his PhD, he has significantly developed his experience in protein biochemistry and mass spectrometry based proteomics as well as bioinformatics and iPSC derived retinal models through his secondments.
I am currently finalizing my project and anticipate submitting my thesis by the end of October, with the defense to follow shortly thereafter. I am actively seeking postdoctoral positions that will enable me to further explore the role of cilia, particularly in the context of various diseases.
What does Shibu say about our program?
I am deeply grateful to the PhD training program and the SCilS Network for providing me with such a transformative experience. The program has been incredibly enriching, allowing me to explore a wide range of methods and techniques in the field. I have also been fortunate to be part of the welcoming and collaborative cilia community, where knowledge-sharing and mutual learning are highly valued. The SCilS PhD group has been the ideal support network throughout this journey, offering encouragement and assistance whenever needed. This experience has been both fulfilling and rewarding, leaving me with memories that I will cherish for a lifetime.
Abstract
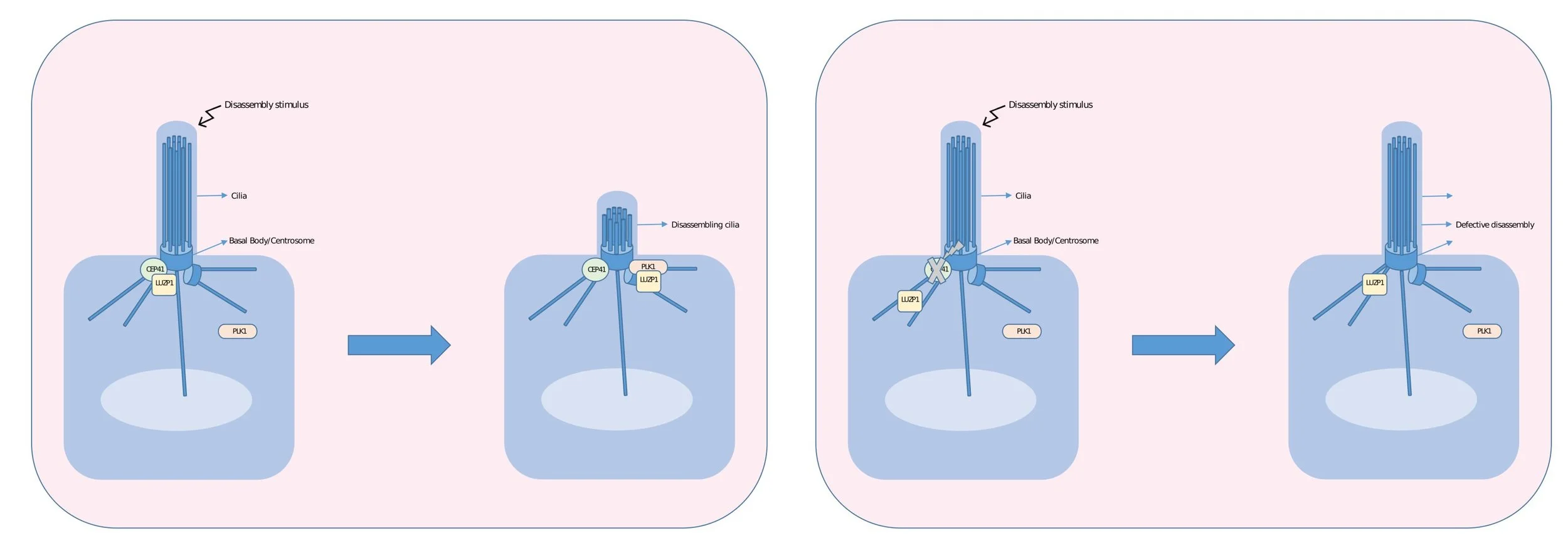
Cilia disassembly is a crucial process for cells to enter the cell cycle and proliferate but also for the function of certain cell types (i.e. retinal pigment epithelial cells). In contrast to ciliary assembly, disassembly is only partially understood and especially the timing of disassembly induction and execution is unclear. In this study, we aim at identifying the protein networks, involved in ciliary disassembly in a time-resolved fashion to understand both the mechanisms of initiation and execution of this process. We will use affinity- and proximity-based methods to quantitatively study the network by analysing the protein complexes of key players and by defining the protein repertoire of cilia at different stages. CRISPR/Cas9 will be used to endogenously tag bait proteins, TurboID, as well as FLAG-based affinity enrichment, will be used to quantitatively analyse the complexes using high-resolution mass spectrometry and statistical analysis of the quantitative data. The initial results will be complemented by alternative proteomic approaches and by localization studies using superresolution microscopy (gSTED) and live-cell imaging. The data will be integrated into the existing ciliary landscape and analysed to define testable hypothesis that will be validated in differentiated retinal pigment epithelial cells.
We want you to understand!
Layman abstract
Cilia are long hair-like structures on the surface of most cells. Cilia are assembled and disassembled during cell division. The mechanisms causing the breakdown of the cilia prior to cell division is something that is not understood that well. My project aims to use different techniques to determine the mechanisms that can explain the timing and execution of this process. The data will be used to better understand disorders caused by caused the dysfunction of cilia.