Ciliopathy disease modules in the ciliary protein landscape
Magdalena Georgieva (ESR13)
Magdalena is from Bulgaria but she shares the MCA principle of expanding her horizons and broadening her expertise through mobility across disciplines and borders. She studied Mathematics and Statistics for her Bachelor’s and integrated Master’s degree in University of Oxford. Developing an interest in biological applications early on, she won research scholarships two years in a row for summer visits to the Dutch Institute of Public Health (RIVM, 2018) and the French institute of Public Health (INSERM, 2019) where she gained hands-on research experience in computational methods in biology. After her Master’s she joined the Protein Evolution lab in Heidelberg University led by Prof. Rob Russell. During her PhD research funded by the SCilS program, she worked on WeSA, a filtering statistic for biological experiments. The program allowed her to work on it jointly with scientists across fields visiting specialists in affinity purification research in Tübingen and structural biologists in Aarhus, while also providing opportunities to exchange with academics more broadly at several research conferences including RECOMB 2022 and Cilia 2022.
She obtained her Doctorate in Natural sciences from the University of Heidelberg in October 2023 and is currently working on crossing borders more literally as a data scientist in the aviation loyalty sector, (Manager Advanced Analytics, Miles & More GmbH).
What does Magi say about our program?
SCilS was an absolutely fantastic program. I really appreciated the meetings we had all together in which I got to learn from colleagues working on the same topic but from 14+ different angles. Both internal project presentations and courses tailored to our SCilS topic were extremely enriching and gave me much more context and opportunity to think beyond my very specific project. And beyond professional advantages, this network fostered relationships with the other fellows which were an invaluable source of inspiration, support and overall fulfilment. The other highlight which I want to share is the mobility support. This, for me, manifested itself in two ways: the internal SCilS meetings and secondments, and the conference funding making intensive exchange within the academic community accessible for us ESRs. I got a lot of experience presenting, tailoring my message to different audiences and listening to others, learning to understand their needs and finding collaboration possibilities - qualities which I find crucial both in academia and beyond.
Abstract
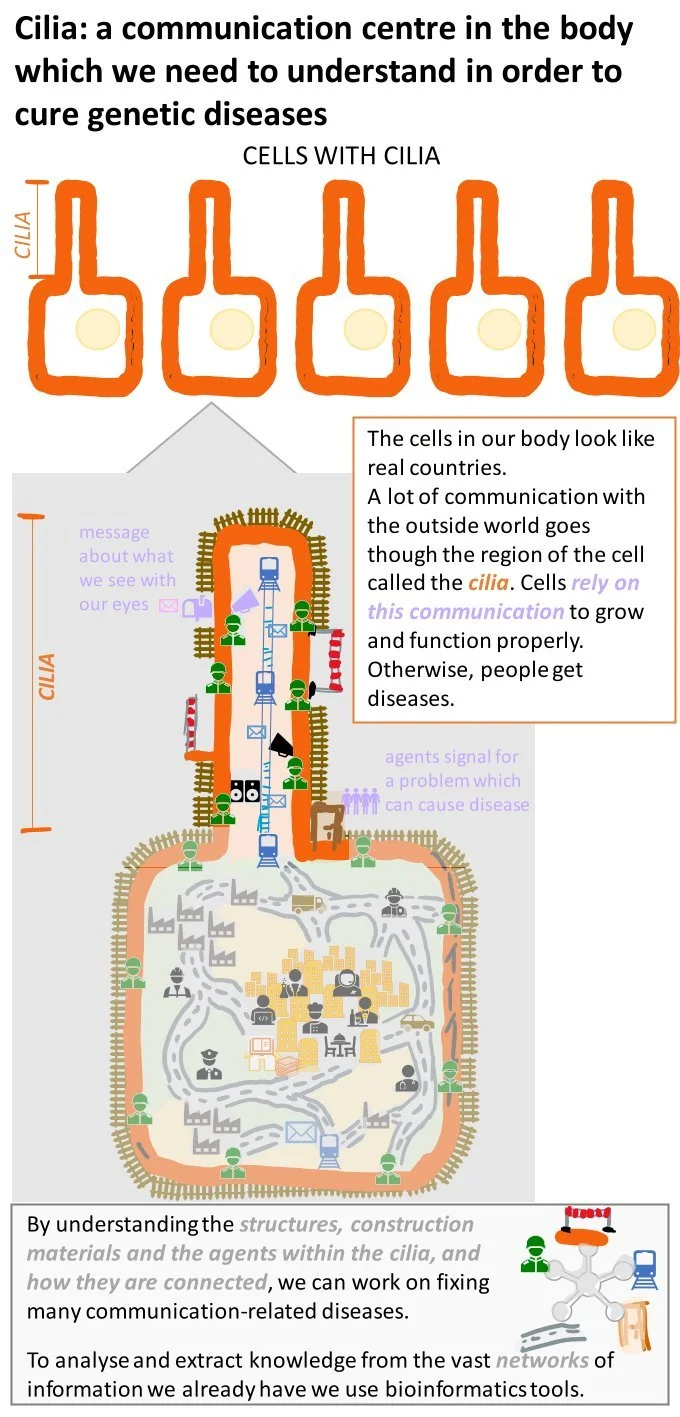
This project aims to integrate the knowledge gathered through new practical research using more precise and advanced methods in structural biology, quantitative proteomics, cell biology, biochemistry and new technologies. In particular, it will scrutinize scilia and genetic variations within it which are expressed as a variety of observed phonotypical and hidden changes. Magdalena’s role in the context of the project is to look at the gathered data from a bioinformatics perspective and apply modelling and statistical techniques to identify patterns and therapeutic targets. All previously available data on ciliopathy mutations in aggregate with new experiments which have been completed as part of this ITN network will provide a rich database for statistical analysis. Expertise within the Protein Evolution group of Prof. Russell in terms of analytical techniques and tools such as the internally-developed socioaffinity method and Mechismo system provide the stepping stone for this bioinformatics project. Those methods will be adapted and used alongside the increasing knowledge of ciliopathy disease genes and mutations with the final aim of creating a schema of all possible complexes and interactions relevant to ciliary disease.
We want you to understand!
Layman abstract
What does it mean for the proteins in our body to work correctly together?
We present a new scoring system, which can be updated as we receive more experiments. It can be applied to many biological networks and helps researchers everywhere to work together and get better results. With this score we do three main things:
• we confirm previous discoveries and find new links between proteins
• we reduce the need of repetitions and control groups which can slash costs for experiments by half
• we can compare the work of mutated and non-mutated proteins and find the causes of genetic diseases.